Program goals
A priority program on “Robust coupling of continuum biomechanical in silico models for active biological systems as a precursor to clinical applications – co-design of modeling, numerics and usability”
Motivation
The great potential of in silico (computer-aided) models is still little used in medicine. As in other high-tech fields, this potential could be used more intensively to accelerate the development of modern diagnostic and individualized treatment methods. Most existing in silico models are limited to single scales (e.g. cell, tissue) or generic multi-scale models of single organs (e.g. muscle, liver). However, in order to understand symptoms and diseases, several size and time scales must be considered in the context of the entire system. The complex cross-scale relationships are often very difficult to identify without a systematic approach and computer models. A central task of numerical biomechanics and biomedical research is therefore to develop robust coupling methods and strategies. These must integrate the scales of biological systems from the molecule to the complete organ system or organism.
Challenge
The high complexity of active biological systems therefore requires close cooperation between medicine, engineering, numerical mathematics (numerics) and computer science. In particular, the description of multi-scale system models requires innovative coupling strategies that incorporate state-of-the-art computer architectures, new and robust numerical methods as well as data structures and integration options. In addition, the simulation results must be prepared in collaboration with medical professionals for transfer to the clinic and for application with clinical questions.
Approach
The proposed SPP will create an interdisciplinary network focusing on research into new methodological approaches for coupling several in silico models and taking into account their physiological functions and three-dimensional organization. Jointly, questions will be defined and addressed that cover the areas of coupling (of in silico models), numerics (robust and efficient algorithms) and usability (standardization and data integration, validation and preparation of numerical continuum models for clinical applications).
Objective
The aim of the SPP is to further develop the existing methodological foundations as key qualifications and thus enable the generation of robust biomechanical models for use in clinical practice. However, the SPP does not aim to establish the transfer of the models into clinical practice via clinical studies. The SPP thus sees itself as a methodologically oriented qualification of biomechanical simulation models of active systems for later use in medical issues. Through the proposed SPP, the existing competencies in the German research landscape can be strengthened in a sustainable and internationally visible manner, resulting in a pioneering role in the field of continuum biomechanical modeling of active biological systems.
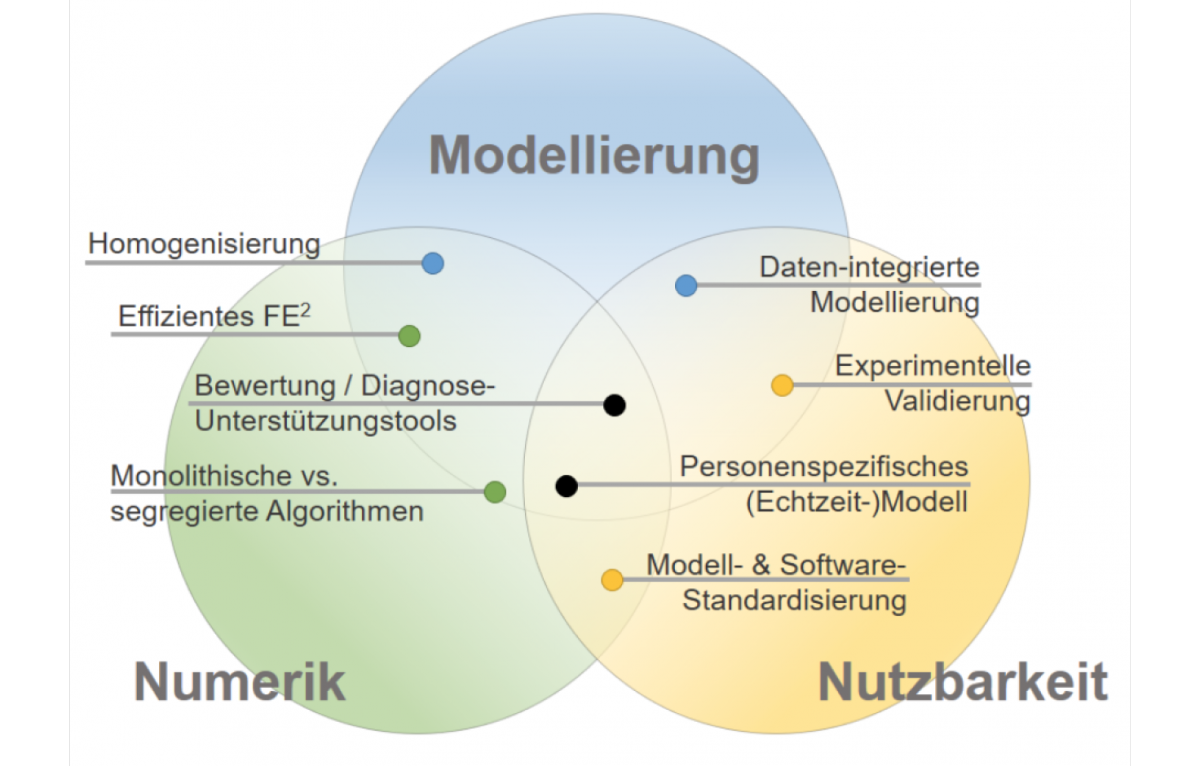
The SPP is divided into three closely interlinked areas.
- The area “Modeling of coupled biomechanical systems” deals with the coupling of biomechanical in silico models for the description of active biological systems on different length, time and/or functional scales or with the coupling of continuum biomechanical organ system models.
- “Numerics of Coupled Biomechanical Systems” deals with mathematical algorithms for the robust, numerical solution of coupled models for active biological systems and their implementation on high performance and supercomputers.
- The “Usability” area deals with data exchange, validation and preparation of numerical continuum biomechanical models to answer clinically relevant questions and for person-specific solutions.
Area “Modeling of coupled biomechanical systems”
In many cases, biomechanical models are initially developed on specific length and time scales that are based on function or structure. As already described, models that span scales and thus the coupling of different scales are necessary to answer clinically relevant questions.
Therefore, the central challenge in the field of “Modeling coupled biomechanical systems” lies in the modeling and mathematical description of different coupling approaches. New coupling strategy approaches are necessary, for example, for bridging scale jumps by coupling detailed models of neighboring scales, e.g. by incorporating the tissue scale, which is usually ignored in multi-scale models of organs, the coupling of two or more functional units on the same scale, e.g. several muscles to a musculoskeletal system or the coupling of two three-dimensional high-fidelity organ models to a system.
Modeling coupling methods that play a role within the SPP are e.g:
- Diffusion-advection reaction equations (PDE /ODE coupling),
- Metabolic processes (systems biology approaches, pharmacokinetic models),
- Transport (blood, hemodynamics – CFD),
- Homogenization methods (mixture theory, theory of porous media, Biot theory),
- geometric coupling (3D-1D-0D coupling) or
- data-driven coupling (e.g. machine learning as a coupling tool).
Examples of spatial and temporal scales on which the integration of different biophysical processes must take place are
- the cellular level (systems biology approaches),
- the tissue level (porous structures, multi-component structures (fibers, matrix, nutrients, ions), multiphases (solids, fluids), perfusion processes),
- the organ level (multi-scale, multi-physical models of individual organs, e.g. the heart, a dissected muscle, the liver, growth process, flow processes), the organism level (coupling of heart and artery, several muscles, liver and kidney) or
- the linking of different time scales (coupling of growth or degeneration processes with transport phenomena of nutrients and transformation substances).
Area “Numerics of coupled biomechanical systems”
When creating biomechanical models, the focus has traditionally been on modeling. Efficient implementation using the latest algorithms, programming standards and existing software libraries is often neglected. However, predictive numerical simulations can only be achieved if modeling and numerics are closely interlinked through strong co-design. It is also necessary to achieve rapid convergence of numerical solution methods for high-fidelity models.
A central challenge for the field of “Numerics of coupled biomechanical systems” is the development of robust coupling algorithms for the different coupled problem classes, e.g. PDE-PDE, PDE-ODE, ODE-ODE, etc., different multiscale problems with different spatial and / or temporal scales or couplings over different dimensions, e.g. 0D, 1D, 2D, 3D, for example to define suitable, non-reflective boundary conditions. A further challenge is to achieve parallel scalability of such algorithms for complex problems on modern high-performance computers. To this end, the algorithms should be developed directly in such a way that they make efficient use of modern computer architectures. As the coupled biomechanical models will generally be non-linear, the development of suitable numerical coupling algorithms must be combined with current algorithms from the field of non-linear solution methods, as otherwise it is often not possible to achieve convergence. A major challenge here is to achieve a balance between the strength of the coupling (monolithic vs. segregated) and the numerical and parallel efficiency, as well as the robustness of the algorithms under consideration. Numerical robustness plays a decisive role for parameter studies, adaptations to person-specific (material) behavior and the transfer to clinical application.
Methodological aspects that play a role within this area are
- Coupling algorithms (monolithic, segregated, …),
- Efficient and parallel scaling algorithms for complex multi-scale models on high-performance and supercomputers,
- Nonlinear/linear solvers (preconditioning, domain decomposition and multigrid methods in space and time, continuity methods, nonlinear elimination methods / domain decomposition methods, …), as far as relevant for coupling and homogenization algorithms, as well as their efficiency and robustness,
- Model reduction (POD, DEIM, neural networks, thin grids, …),
- Numerical homogenization methods (FE2, …) and
- Operator splitting methods.
Area “Usability”
Biomechanical in silico models have the potential to quantify certain complex processes and thus, for example, predict or enable the benefit and detailed planning of surgical interventions or support their implementation in the operating room. They can also help to improve scoring systems, such as the established LiMAx evaluation system. The necessary quality assurance of computer-aided simulations requires the analysis of the sensitivity of individual input parameters to the final result with regard to reliable predictions.
One of the biggest challenges in the area of “usability” is the creation, exchange and provision of validated models and the integration of patient-specific information. In addition to data integration, the link between ex vivo and in vivo data plays a central role in achieving new insights and improving existing models. In contrast, the in-house development of software to solve biomechanical problems usually leads to isolated solutions that make the exchange of models and data more difficult and prevent new innovations.
By integrating data from various resources and using them intelligently, e.g. through machine learning, the reliability and informative value of in silico models can be increased enormously. In addition, validation studies must be designed so that the functional reliability of complex simulation models can be convincingly demonstrated and user acceptance increased. The robustness of the in silico models should be analyzed and experimentally verified as part of clinically relevant sensitivity studies. Furthermore, quantified analysis with the aid of in silico models presupposes that the corresponding tools can also be used by medical professionals without a broad technical background. The underlying models must therefore be very robust and equipped with an intuitive and generally understandable user interface. In addition, methods must be found that allow morphological and mechanical parameters to be extracted from imaging procedures and incorporated into robust three-dimensional in silico models.
Methodological aspects that play a role within this area are
- Validation with
- Co-design of experimental methods,
- Statistically validated validation methods for coupled biomechanical systems,
- Generating new data for the overall system,
- Simulation-based analysis and interpretation including in vivo / ex vivo link,
- Standardization with
- Data standardization and data management, e.g. through the further development of existing XML standards,
- Integration of data from various resources and their intelligent use and
- Application with
- Data processing,
- Real-time capable systems, e.g. with the help of machine learning / neural networks,
- Definition and validation of scoring systems, e.g. for therapy or implant selection through a data simulation analysis / interpretation cycle,
- Creation of the workflow in a clinical environment, e.g. surgical planning software, diagnostic software and intuitive user interfaces and
- Reflection studies to increase social / internal medical acceptance
- of simulation models in translation to clinical applications.
Thematic restriction (positive list)
In order to narrow down the topics, only research activities in which coupling methods play a central role are to be funded in the SPP. Furthermore, only the following topics should be explicitly approved for application.
- For the area of modeling coupled biomechanical systems, the active biomechanical systems considered in the application should deal with the following properties:
- Soft tissue,
- Multiphase materials / multiphysical models,
- Active materials, e.g. force, metabolism or signal processing and
- Continuum mechanical models.
- For the area of numerics of coupled biomechanical systems, the methods should have the following characteristics:
- Coupling algorithms for the solution of coupled systems,
- Algorithms for scale bridging and / or homogenization, e.g. FE2 and
- Increased efficiency, e.g. model reduction, HPC (parallelization, data communication, etc.).
- The following characteristics should be taken into account in the area of usability:
- Multimodal data and data integration,
- Standardization, validity, quality assurance or data management and
- Relevant medical issues.
Thematic restriction (negative list)
Explicitly excluded are
- passive biological structures if only mechanical aspects are dealt with,
- the further development of single-scale or single-phase models,
- purely dynamic rigid body models,
- in the first phase of the SPP projects on uncertainty modeling,
- Research that focuses purely on subcellular processes (-omics, DNA, mRNA, proteins, ion channels),
- clinical studies and
- experimental work that is not specifically related to modeling.
Although the above-mentioned aspects are also important for the issues addressed by the SPP, they cannot be included due to the necessary focus and desired coherence.
Application formats
Applications should be thematically anchored in at least two of the three areas. The aim is to divide the projects into
- up to 10 applications with 24 person-months per year and
- up to 5 applications with up to 36 person-months per year.
Examples